Types of Genetic Testing Utilized for Personalized Medicine
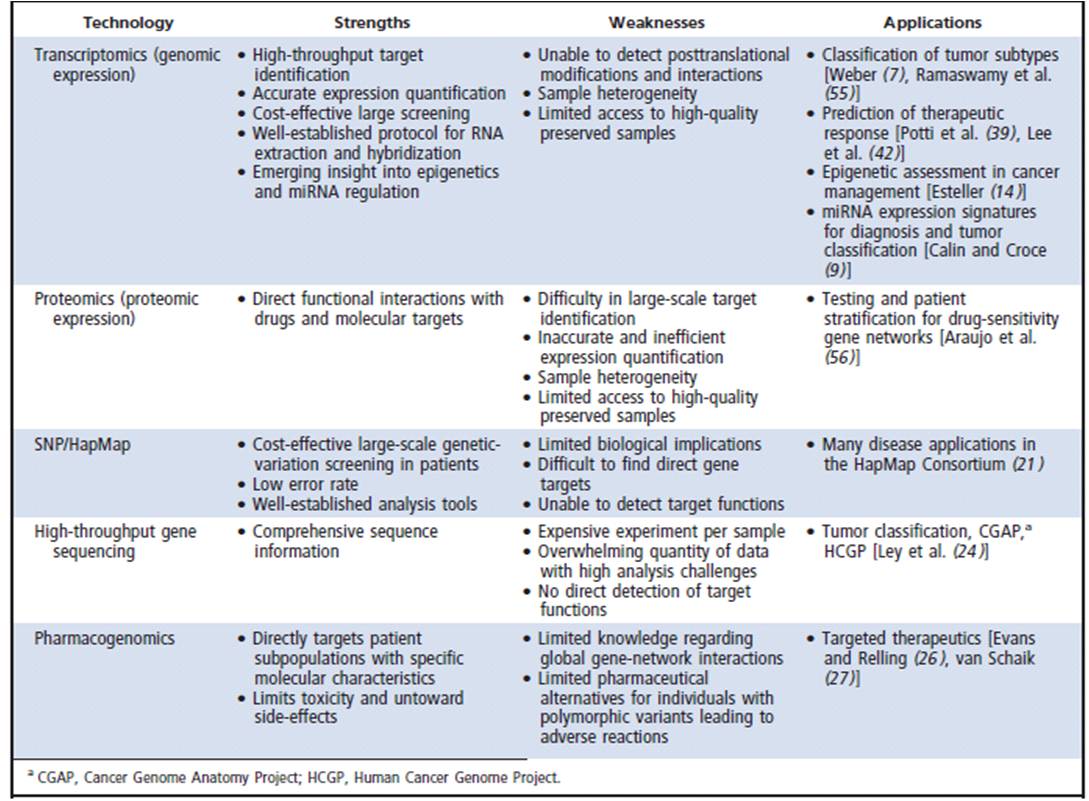
The table above compares advantages and disadvantages of different techniques for genetic testing. Taken from Overdevest et al. (see References)
Transcriptomics
This form of genetic testing aims to gather information about the gene expression levels of many different genes through the use of microarrays. Although many individuals may carry a gene for a particular type of cancer, the person will not develop the disease until those genes are transcribed and then translated to make protein products. Thus, to determine an individual's susceptibility to developing a particular type of cancer, the expression levels of these genes may be tested. To determine this, mRNA is isolated from a patient's tissue sample (removed either through surgery or from a biopsy). From this mRNA, researchers can create the more stable complementary DNA (cDNA), which is fluorescently-labelled. Short segments of this cDNA are then applied to an array where mRNA segments, representing different genes, have been immobilized. If cDNA segments find their complement on the array, they bind at that spot. After an incubation period, unbound cDNA segments are washed away, leaving only those segments bound to mRNA on the array. Then, the amount of gene expression can be determined by measuring the fluorescence of each spot on the array, representing an individual gene. The DNA microarray presents a robust platform for high density testing of gene expression levels.
The video clip above illustrates the process of genetic testing using a DNA microarray.
In addition to detecting segments of mRNA representing the expression of regular genes, DNA microarrays have also been adapted to identify microRNAs, which are noncoding sequences about 20 base pairs long which act as either oncogenes (which promote the formation of tumors and the development of cancer) or tumor-suppressor genes. The presence of these microRNA segments can provide more information about the progression or development of the disease.
[back to top]
Proteomics
In proteomic analysis, microarrays are also used. Instead of measuring the gene expression levels, however, the abundance of the products of these genes (i.e. their proteins) is detected. Here, a microarray is composed of either antibodies or antigens, which can bind antigens or antibodies, respectively, in a patient sample to indicate how much of a particular protein or antibody is present. The benefit of proteomics over transcriptomics is that proteomics allows researchers to probe the interactions between proteins, which provides different information than the gene expression levels alone. Though this technique may provide a broader view of complicated interactions, the number of protein products far exceeds the number of genes. Due to this fact, transcriptomics may be used for initial studies of gene expression, with subsequent proteomic studies that focus on a smaller set of proteins.
[back to top]
Haplotype Mapping
Although the genetic sequence that comprises the human genome is widely conserved from one individual to another, small differences do exist. These small differences, occuring when a nucleotide at a certain position in one person's genome differs from the nucleotide at the same position in another person's genome, are termed single nucleotide polymorphisms (SNPs). A haplotype comprises many of these SNPs from a single person. The concept of haplotype mapping is that certain haplotypes will occur more often in populations of people who become afflicted with a certain disease, while other haplotypes correspond to individuals who are disease free. Then, a person's proclivity toward disease, or response to a certain type of treatment, can be predicted based upon that person's haplotype. Haplotype mapping provides a quicker way to identify possible biomarkers for disease than whole genome sequencing. Since SNPs provide only binary information, however, a large number of them must be used to achieve accuracy using this technique. In addition, haplotype mapping has no way of measuring the post-transcriptional modifications that occur, which can contribute strongly toward the development of cancer.
The clip above describes single nucleotide polymorphisms (SNPs).
[back to top]
High Throughput Genetic Sequencing
The whole genome may be sequenced using several different methods. The clip below illustrates Sanger sequencing, an older technique that was used for the Human Genome Project.
Since that time, higher-throughput methods of sequencing have developed, including the use of microfluidic devices (Paegel et al., see References page). Several companies, including Illumina and Roche, have also developed high-throughput sequencing methods. While these methods have enabled faster sequencing at lower cost, whole genome sequencing fails to detect post-transcriptional modifications, which can have an important effect on disease development.
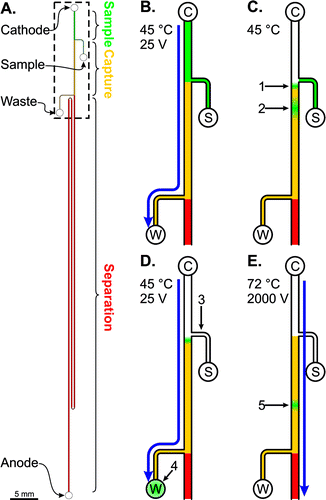
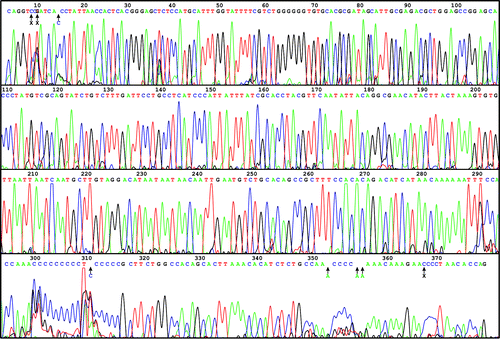
A microfluidic device for Sanger sequencing, and the fluorescent readout from the device. Blazej et al. 2007
back to topPharmacogenomics
This technique aims to identify polymorphisms in genes which will guide the selection of a pharmaceutical agent for treatment. Specifically, sites which encode factors involved in metabolism, or potential receptors for drugs, are analyzed. However, this technique will be imperfect until researchers understand all of the possible polymorphisms for nucleotides and what they represent in each case.
back to top